Using Deep Learning to Combat Antibiotic Resistance
Can a computer identify bacterial genes that code for antibiotic resistance? If so, can it also tell us to which antibiotics they are resistant?
Working on these problems in Iowa State University College of Veterinary Medicine are Md Nafiz Hamid, a PhD candidate in the Bioinformatics and Computational Biology Program, and Iddo Friedberg, an associate professor in the Department of Veterinary Microbiology and Preventive Medicine. They are using cutting edge computational methods, known collectively as deep learning, to train computers to mine bacterial genomes for genes associated with antibiotic resistance.
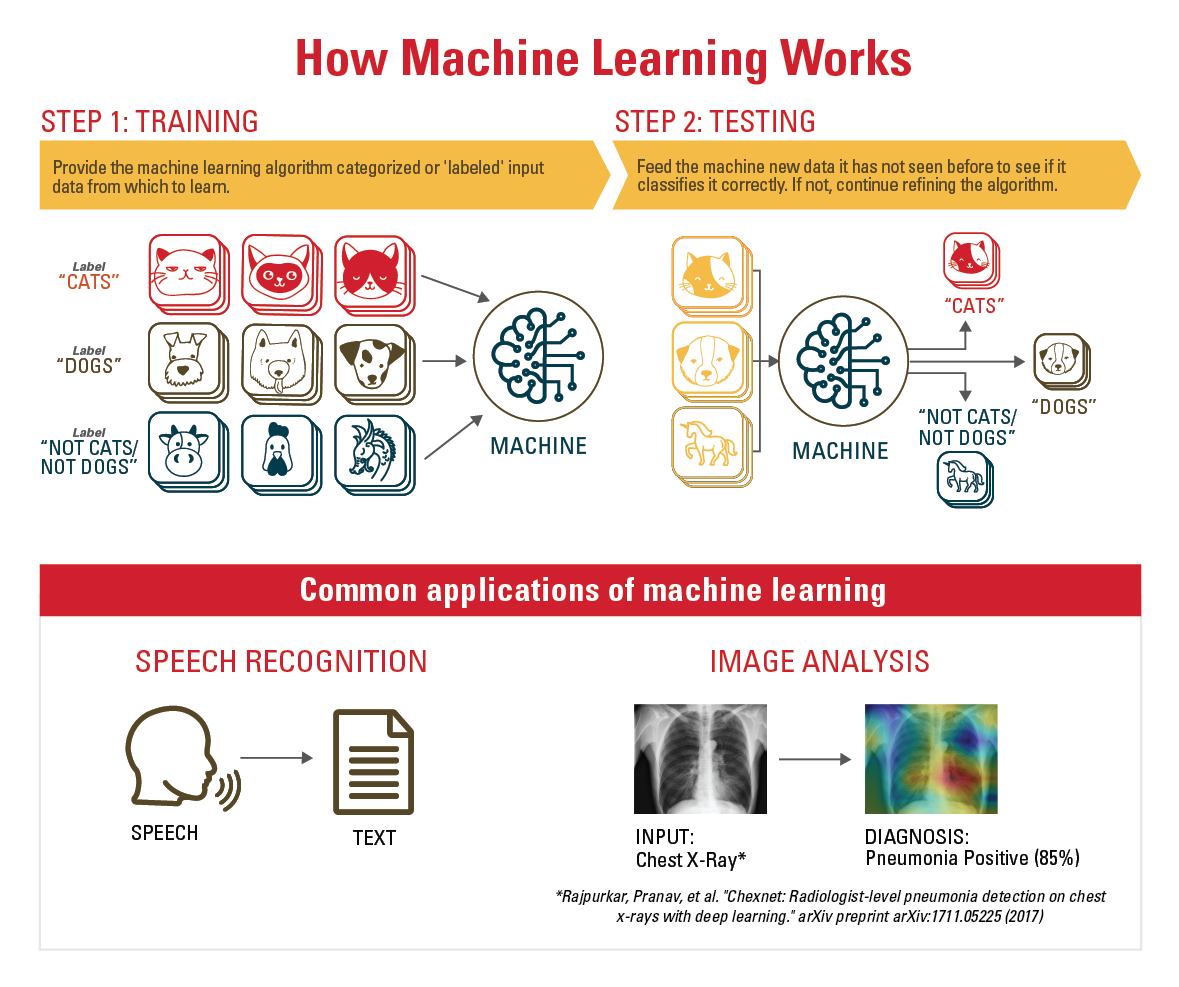
Deep learning is used in a wide variety of applications: self-driving cars, speech recognition (Siri uses deep learning), and handwriting recognition, to name a few. Recently, deep learning programs have been shown to provide highly accurate predictions of the likelihood of melanoma or cervical cancer. “The programs were trained on images typically shown to physicians for diagnosis,” Friedberg says. “The accuracy of the trained programs, once they were shown images they had not seen previously, was better than that of human experts.”
How did they do it?
Hamid wrote a program that reads the sequences of genes that provide antibiotic resistance to each of 15 different drugs. His program learned the patterns that are typical for each type of resistance. To train the program, he used thousands of sequences of proteins that were verified to be involved in antibiotic resistance to one of these 15 drugs.
“But, in the real world, deep learning models will encounter proteins that are resistant to antibiotics other than the 15 they were trained for, or, most commonly, proteins that may not have anything to do with antibiotic resistance at all,” says Hamid. “Even if a bacterial protein is not identified as being involved in antibiotic resistance, it may still have that yet-undiscovered function,” he said. “Deep learning methods may still wrongly classify these proteins.” This misclassification error is a known problem in many deep learning applications.
To reduce this type of false positive error, Hamid introduced some noise — deliberate errors — into the learning process. By doing so, the program had to overcome more hurdles while learning: it worked harder to identify antibiotic resistance genes, and as a result it actually became more accurate in its predictions. Hamid then tested his program on a set of genes that the program never “saw” before, and his model was highly accurate in predicting the antibiotic resistant genes, with a lower false positive rate than similar programs not trained using Hamid’s noisy method. Even when the program was presented with negative data such as human genes that are not expected to have antibiotic resistance, or bacterial ‘housekeeping genes’, it could identify these sequences as having no antibiotic resistance.
Hamid’s work was accepted for presentation in the Machine Learning for Health workshop in the prestigious Neural Information Processing Systems conference, the premier international conference on machine learning.
This is not Hamid’s first foray into using deep learning in mining bacterial genomes: he and Friedberg recently published a paper on using deep learning to discover bacteriocins: short antibacterial peptides that are candidates for many different applications, from drugs to food preservatives. To do so, Hamid used a technique developed by Google used to classify web pages, and adapted it to better identify bacteriocin-coding genes in bacterial genomes.
#
Md Nafiz Hamid is a PhD candidate in the Bioinformatics and Computational Biology (BCB) graduate program, where students are trained at the interface of computer science and biology. Iddo Friedberg, PhD, is an associate professor in the Department of Veterinary Microbiology and Preventive Medicine, and will become the director of graduate education for the BCB program this summer.
December 2018
Further reading:
[1] Hamid MN and Friedberg I Reliable uncertainty estimate for antibiotic resistance classification with Stochastic Gradient Langevin Dynamics (2018) Machine Learning for Health (ML4H) Workshop at NeurIPS 2018 https://arxiv.org/abs/1811.11145v1
[2] Hamid MN and Friedberg I Identifying antimicrobial peptides using word embedding with deep recurrent neural networks (2018) Bioinformatics bty937
[3] Wainberg M, Merico D, DeLong A, Frey BG Deep learning in biomedicine Nature Biotechnology (2018) 36:829 https://www.nature.com/articles/nbt.4233
